Quick reminder! You can get my posts fresh in your inbox if you join the mailing list here
I'm telling a little bit of a lie here really. When I say fish, I really mean poisson and when I say poisson I really mean Poisson Disc Sampling. Turns out, Poisson disc sampling is really useful in my A.I research. Problem is, it's not the fastest process in the world, particularly when python is involved. So what can we do? Turns out there are lots of options - investigating them has been fun and might be useful for other folks working in this area.
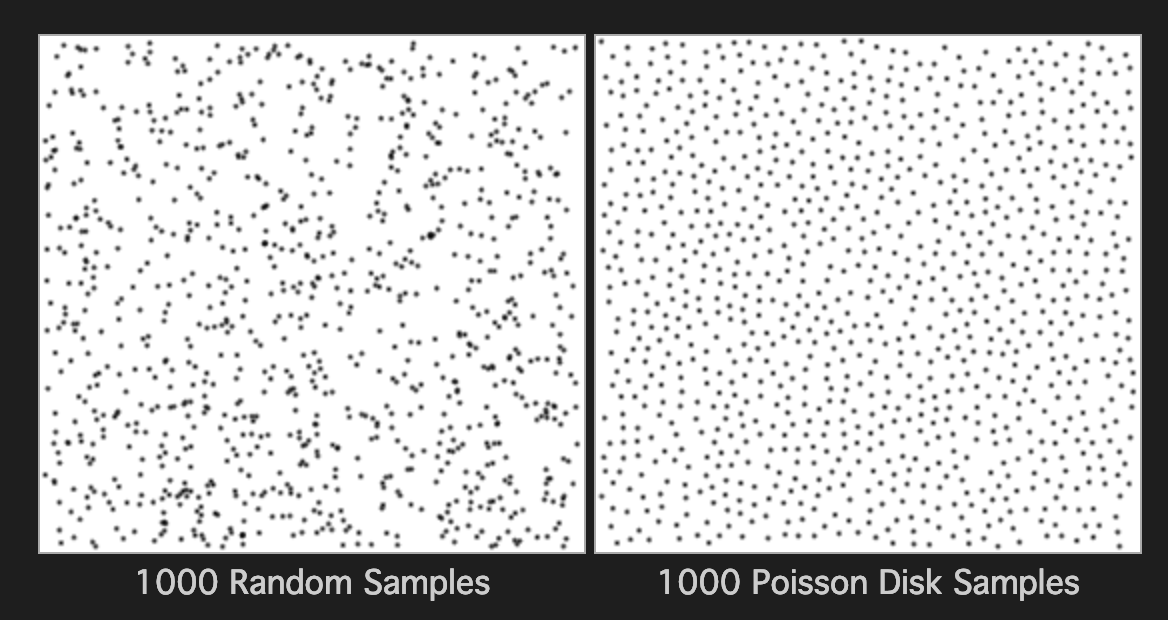
Poisson Disc sampling is a method for creating pleasing random patterns, or rather, random distributions with a somewhat regular spacing. Take a look at the image below and you'll see what I mean.

I believe the name comes from the Poisson Distribution - I'm not really sure to be honest. How I understand the algorithm is you start with one point and then add another within a circle or sphere of a set distance away with an increasing probability towards the boundary. The idea is to maintain a minimum distance from one point to any other point. This leads to a bit of a problem though as it's quite computationally expensive to check each additional point against all other points. There have been a few attempts to speed things up. I settled on Sample Elimination for Generating Poisson Disk Sample Sets algorithm by Cem Yuksel. This algorithm works by creating a large random set first, then removing points from the set until the Poisson property is - more or less - obtained. It relies on two data-structures - the kdtree and the heap.
Sample elimination assigns weights to each point, adding them to the heap as we go. We then take a point off the heap, finding all these points nearest to this point using the tree. With this point removed, we rebuild the heap using the weight formula, and go again, until we arrive at the number of point we want. In my tests, one needs roughly 8 time the number of points you want in order to get a good final result.
I'm a tad bored of Python; it tends to dominate the AI world at the moment. I believe there are a few other languages people can use but the vast majority of code seems to be in Python. It has some advantages - it's fairly easy to get into and works on most platforms, but like all interpreted languages, there is a bit of an overhead. Python performance has improved over the years but it still isn't top. However, the first place to start in speeding up your code is to take a look at how you've built the algorithm.
Firstly, I removed many of the sqrt() calls, as we merely want to order the points in the sample rather than record the actual distances. This seemed to have some effect on performance though not as much as I'd thought.
One thing we can do is measure where our code is taking the most time. Python has a number of profilers such as cprofile and vprof to name a couple. They are quite easy to use too. Once I measured where my program was spending it's time, I decided I needed to remove numpy from the equation and work with some simpler datatypes, reducing any unnecessary conversions. Around this time, I was made aware of a package called numba.
I thought I'd ask around to see what other people were doing to speed up their scientific code. What better place to start than the UK Research Software Engineering group. I quickly received the answer I was looking for - some software called numba.
Numba is a JIT -Just in Time Compiler that aims to speedup certain python functions. Certain parts of python and numpy can be compiled down into a faster code blob. One need only decorate the function in question and replace a few types and Boom! - faster code.
In practice, it's a little more tricky than that. One needs to do a little conversion from complicated to more simple types that numba can handle, but this isn't too onerous.
I've spoken before about Rust and how I enjoy writing code with it. I spotted an article recently about how scientists are moving to rust. Rust's popularity among programmers is quite well known. I think this is partially because it's a hard language to learn initially and requires some knowledge of the machine, data-types, borrowing and the like. It certainly strokes your programmer ego, which isn't such a bad thing if kept in check. I certainly feel a sense of accomplishment when I write working code in Rust - something I never really get in Python. It definitely takes longer to write Rust code than Python, but it feels better and runs faster. The code seems tighter, leaner with fewer bugs and more tests (I think tests are easier to write in Rust than in any other language). It's become my goto for writing any performant code.
I rewrote the algorithm for Poisson Disc Sampling in Rust and received a reasonable amount of speedup over numba. Not a lot mind you, which was a surprise, but enough to keep going and see how far I could push it.
One of the easy wins for speed is to parallelise your code. In Python, I've always found this to be more difficult than it should be. I also think Rust could do better in this regard. In the past, Rust used to have the scoped keyword that helped in determining how long variables would live for in each individual thread. This was removed some time ago however, which was a bit of a pain. Now, if we want to do work with threads, it's somewhat tricky - even the simple pattern of fork-join. Not exactly living up to Rust's touted 'fearless concurrency' I think.
Fork-Join, or divide and conquer is a really simple approach to parallelising your code. Divide up the dataset, work on each bit individually and once all the threads are done, combine the results together. Simple right? Well, perhaps not as simple as you might think.
Rust's main strength and it's main barrier to understanding is the borrowing mechanism it works on. Variables have lifetimes and are owned by parts of the program. Threads and concurrency mess with this sort of thing a lot. I mean, can you guarantee that a thread will stop before the code that called it ends? What happens to a shared variable, particularly something like a vector that can be modified at any time by multiple processes? It is a tricky thing to have to write yourself, made more difficult as there are a lot of options in Rust, many of which are not built in as standard.
Having written the fork-join algorithm a couple of times, I've found a way that I think works quite well. I used the scoped-threadpool crate and some mutexes. I've tried things like Cells, Arcs and a whole manner of oddly named libraries but this method seems to be the simplest:
let rsamples :Mutex<Vec<Quat>> = Mutex::new(vec!());
let rpoints : Mutex<Vec<Quat>> = Mutex::new(vec!());
let mut pool = Pool::new(partitions as u32);
pool.scoped(|scoped| {
for i in 0..partitions {
let bsamples = &rsamples;
let bpoints = &rpoints;
scoped.execute( move || {
let ns : usize = sample_size / partitions;
let mut tpoints : Vec<Quat> = vec!();
for p in points.iter() {
let aa = p.axis_angle();
// DO ALL THE WORK HERE
}
//println!("{}, {}", i, tpoints.len() );
let trval = sample(&tpoints, ns);
for q in trval.0.iter() {
let mut t = bsamples.lock().unwrap();
t.push(*q);
}
for q in trval.1.iter() {
let mut t = bpoints.lock().unwrap();
t.push(*q);
}
});
}
scoped.join_all();
});
(rsamples.into_inner().unwrap(), rpoints.into_inner().unwrap())
Like so many things in life, when you figure it out, it becomes quite simple. There are many crates out there that provide fancy options for Rust concurrency but scoped-threadpool is simple enough. We create a pool first, then call scoped.execute as many times for as many threads as we want, using the move keyword to move our variables into the scope of each thread. The key here is we are moving in a reference to our shared vectors at the top of the code, so each thread can access the vectors. By wrapping the vector in a mutex we guarantee it can only be accessed by one thread at a time. This is quite similar to how we tend to write this sort of algorithm in languages like C++. Sometimes, it can be easy to be distracted by the fancy crates and syntax rust offers, but the basics are still there doing a good job.
Parallelising the Poisson Disc algorithm is not without it's problems. Dividing up the space into equal chunks and performing the sampling on each chunk works a bit, but it one isn't careful, you can get some bad artefacts. Consider a chunk where there are a large number of samples - say 1000 - and you only want 2 points. Typically, these points will be close to the border of the space in question, which makes sense if you want to maximise the distance between all the points in your sample. However, when you combine these chunks, you get a pattern where the points all lie along the joins of the areas. Not ideal.
This basically means we can only go so far with parallelising the algorithm to get some speed up. Still, it's better than nothing.
So Rust and threading gets us to good-enough performance. Now how do we get rust to talk to Python? There are a few ways, including writing results to a file and having python read it, but perhaps there's a better way? Asking around on the UKRSE group again, a few folks suggested ctypes, but I decided to go with PyO3. PyO3 works in both directions - either calling Rust from Python or vice-versa. In my case I want to call Rust from Python, sending some simple data in, and getting a load of points back.
In order to do this, we include the crate in our rust program, then write something a little like this
use pyo3::prelude::*;
use pyo3::wrap_pyfunction;
/// Formats the sum of two numbers as string.
#[pyfunction]
fn sum_as_string(a: usize, b: usize) -> PyResult<String> {
Ok((a + b).to_string())
}
/// A Python module implemented in Rust.
#[pymodule]
fn string_sum(py: Python, m: &PyModule) -> PyResult<()> {
m.add_function(wrap_pyfunction!(sum_as_string, m)?)?;
Ok(())
}
Pretty straight-forward really. I place my functions inside a lib.rs file, creating a shared library and a python module I can import into my Python code as normal. Pretty neat!
Well lets do a basic test with Python's unittest module. Lets start with a pool size of 200, reducing to a final sample of 8.
python -m unittest test.poisson.Poisson.test_speed_rust
.
----------------------------------------------------------------------
Ran 1 test in 0.013s
OK
Now, lets go with numba
python -m unittest test.poisson.Poisson.test_speed_numba
.
----------------------------------------------------------------------
Ran 1 test in 1.782s
OK
Looks like Rust is a lot better at first glance but 200 is far too small to be sure. Lets try 8000, reducing to 200. Rust comes in at...
Ran 1 test in 373.959s
Lets see how numba fares...
...
Oh dear. It never actually finished. Well nevermind that then. Don't get me wrong - I think numba is a great idea and I'd rather have it than not. For a quick fix it's great, but if one needs to learn some extra syntax, extra types and so forth, it might be better to learn a new language. I suspect it'll serve you well in the longer run. I could spend the time really getting to know numba - I know I've not used numba to it's fullest - but I think my time could be better spent. Maybe I'll change my mind in the future? I'm still glad to have given it a go.
It turns Poisson sampling my data for each batch improves performance massively, and I mean the not-working to working kind-of massive! It's really turned things around in our particular problem case. Although this increases the time taken to perform a training run, the final results are much more likely to be acceptable. We only need to poisson sample once for each run, so while there is one up-front cost, the training time remains the same. It was definitely worth the time to bring Rust and Python together.
Like any other engineering discipline, in software engineering it pays to use the right tool for the job. In my case, Python and PyTorch have the AI bit covered, but for some nice speed on a straight forward number crunch, there's no betting something like Rust, C++ or even Fortran. The Python folks know this and so there are plenty of Python wrappers to faster, low-level libraries. It all depends on what you need to get done.
All this said, I'm intrigued by Julia. I think that might be next on the list to learn.
