I was fortunate enough to be admitted on the December round of the Alan Turing Institute Data Study Groups at the British Library here in London. It's a 5 day hackathon where you get to work on some big, interesting (and often classified) datasets. The idea is these data are related to real problems that have significant impact.
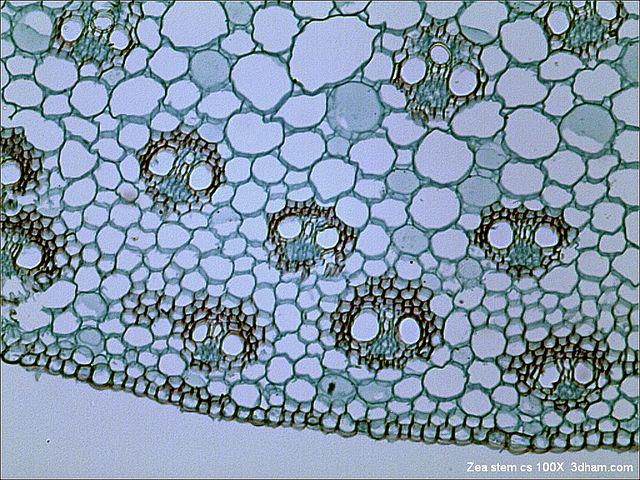
In this round we had projects from DSTL, the WWF and the National Archives among others. I was quite keen on the Bright Field Microscopy project as it's fairly related to what I'm doing at King's College. The idea, in a nutshell, can you identify the cells in the image.
Cell segmentation is quite the challenge - indeed there is a competition to find the best performing algorithm. Deep learning has been used in a few similar cases so our team figured this might be a good place to start. I suggested we try the U-Net algorithm as I figured it might have a chance to at least remove the background, at which point, getting the cells themselves might be easier.

The Alan Turing institute is a bit of an odd duck. It occupies one floor of the British Library, with a couple of rooms dotted about elsewhere. It feels very much like a cross between a startup and an academic outfit. It's a bit like the YouTube HQ offices somewhat I suppose, only a little smaller. The Turing seems to have a mission to be a fixer of sorts - making connections and bringing folks together in teams to have impact.
Yes, that word appeared a few times in the spiel. It crops up a lot in academia and publishing these days. However, I get the impression the Turing really wants to get to these interesting practical problems to see if big-data and machine learning can help.
Our team was one of the largest (the other being the WWF). We were placed around a bunch of desks and told to get cracking. One of the students has been selected as a facilitator and made a good job of setting the framework for how we'd tackle this problem. Essentially we had a little Kanban going, with group meetings scheduled throughout the day.
I found the room we were in to have annoying lighting and not enough room, so I relocated to a hot-desk area further into the building. It was quite nice, though perhaps not ideal for bouncing ideas around. Nevertheless, the window, comfy chairs and endless coffee and tea meant that it wasn't a bad place to get some serious coding done.
I noticed a lot of my teammates were using Jupyter notebook inside a browser inside a VM, training and testing large networks on GPUs. I was somewhat shocked and horrified! This is no way to do engineering! I suspect, given the nature of the challenge, quickly slapping things together however you can is fine, but when someone asked me to help them with their version of U-Net, showing me a half screen of unresponsive text, my heart sank. I should have said no, debugging is painful enough!
The Turing makes a big deal about being able to handle sensitive data. It uses a sort of Sharepoint, Virtual Machine setup, with a web-browser interface. Everything inside the haven is walled off. Cut and paste doesn't work, and there is no internet access within the VM. Needless to say, this makes things incredibly clunky! I found a work-around eventually, but having to use putty inside a vm, inside a browser, on a mac was just the worst!
So in the end, I decided it would be best to find some other dataset to work with that looked similar enough. This isn't a bad idea anyway as having a small dataset to test on is something I'd consider regardless of a data-haven. I found a HeLa cell Bright-field dataset and got cracking!
U-Net is a convolutional neural network architecture, so called because the diagram looks like a 'U'. It's very similar to the encoder/decoder style of network where you shrink down a large datum into a simpler representation, then scale it back up again. The hope is that the network learns the important points in, say, the image, then gives you back an image that is hopefully close to what you want. Image in, image out.
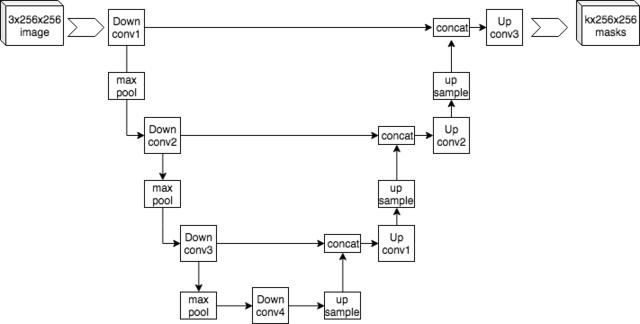
U-Net differs in that it has a set of cross links. As we come back up the U we concatenate out convolutional layers with the output from the corresponding point on the down part of the U. This means you get an interesting mixture of input and output as your output. I'm not sure what the rationale is but I've seen examples that look compelling. Saying what is a car and what isn't inside an image, effectively creating a mask, seems to be no trouble at all for this network. In fact, there is a nice implementation in pytorch on github. I cribbed a bit of it for our challenge.
Cell Segmentation seems to be quite an important topic for biologists. It's a good computer vision problem to wrap your head around, as Cell's are an odd shape, often closely packed, with little contrast. Our group had a few ideas of what to try. In addition to U-Net and data augmentation, we had a look at the following techniques.
Mask R-CNN is one such approach that draws bounding boxes around detected objects. I'm really not sure how it works but the code and PDF are available. From what I saw, it looked like it was working quite well.
Distance Fields are not a solution in of themselves but they could be the key ingredient to making a thing work or not. If I was a betting man, I'd put money on distance-fields-on-borders + U-Net giving good results. A distance transform results in a distance field where you know how far you are from the nearest point of interest. So if I pick a pixel at random, it'll have a number that corresponds to how far that pixel is from the nearest mask. The advantage here is we get a smooth gradient - a clue - that our neural network can work upon.
GANs or Generative Adversarial Networks are all the rage at the moment. They are quite impressive in what they do. The idea is simple but genius. Train a network to produce the image you want, whilst training another network to spot a fake. Each network reinforces each other; one getting better at fakes, the other getting better at spotting them. By the end of the day, one should hopefully see some convincing results. Combined with the distance field approach, GANs seemed to be working quite well on cell segmentation, last I looked.
The real trick and challenge in this, er, challenge is that we don't want to just say what is cell and what is background (which U-Net can do reasonably well). We want to identify the individual cells. U-Net has a useful ability in that it can place a pixel in one of many classes, not just one of a binary set. However, we don't have any specific classes for our cells, just that this is one cell and this is another. Separation is what we are after and that's what make's this a difficult challenge.
I managed to get U-Net to give reasonably good semantic segmentation. This is a fancy word for finding-a-mask-for-background-not-background. From there I hoped to get some sort of segmentation but I didn't have the time. U-Net is pretty good at providing more than just 2 classes though, so if I need to identify specific cells, U-Net is a pretty good shout, but in this case, it's not ideal. When I start to look at the worm neurons for my PhD work, U-Net will come in very handy!
It always pays in spades if you spend time looking at the data and how it is represented. Some of the data provided by DSTL came in a 16bit tiff. It turns out that Python PIL / Pillow has a bit of trouble reading these correctly, so it's definitely worth checking your assumptions.
Later during the week, a few folks converted the 8bit masks we had into distance fields. Feeding these into the GAN approach resulted in distance fields that could then be fed to a watershed like algorithm for final segmentation. Not a bad approach indeed, and made possible by a relatively simple data transformation.
Because of the data-haven annoyance, I decided to look at a public dataset, because there are many out there that aren't so different. I found a set of HeLa cell images from the celltracking challenge website.
One approach I attempted with U-Net was to detect not areas but borders. We can generate borders from masks easily enough but such a dataset has lot's of empty space punctuated by an occasional line of pixels at maximum brightness. Not easy for a neural network to solve. However, using a distance field approach might work well.
Data-augmentation is quite the interesting challenge in-of-itself. I wrote some code of my own to augment the data we had, rotating images in the 4 cardinal directions (90, 180, 270 degrees basically). That way, I avoid artefacts and because the images are square, no scaling or cropping is required.
I couldn't make the last day, on account of having lectures and meetings I didn't want to miss, but each group had to submit a report and give a presentation. I had a bit of bother getting on the same data page as the other folks in our team (largely because I'd already done my dataset sorting really early on) but it seemed alright in the end.

You can find my code at https://github.com/OniDaito/turingdsg. It should run without too much trouble on the HeLa dataset. It needs a little more work but I'll be coming back to it once I've started looking at my worm brains!
Working in a more team, exploratory, scrappy way with jupyter notebooks in a crowded room, with a difficult data-access policy were the worst elements of this week for me I'd say. Having the occasional one-to-one chat with some of the folks there, the tasty snacks (that's how they getcha!) and learning about techniques I'd never heard of before were the real highlights.
I'd say, for someone more social, more data-sciency than engineering and with more time, this is a good thing to try. If there is a challenge on their list that sounds appealing then I'd say apply and give it a go. The application process involves writing a few paragraphs on what you want to get out of the week, your qualifications and background and what not. It takes about 45 minutes but it's not too strenuous. I'd say you need to know your data-science and AI. Very quickly, folks were trying some of the latest techniques from recent papers, It's impressive to see really.
If you are fortunate enough to have access to a good GPU, a place to work and nice colleagues you can bounce ideas off already then the Data Study Group probably won't be of much use. That said, chatting to folks who aren't data-scientists but project owners is useful. These folks know a lot about their particular area and it can range from archivists to conservationists to local councillors. It's interesting and fun to talk to folks outside your area from time to time, especially when you've got a good excuse to work together on a thing. It's a good ice-breaker.
A mixed bag for me personally, but overall I think it's a good idea and I'd encourage folks to apply if this sounds like your bag.
