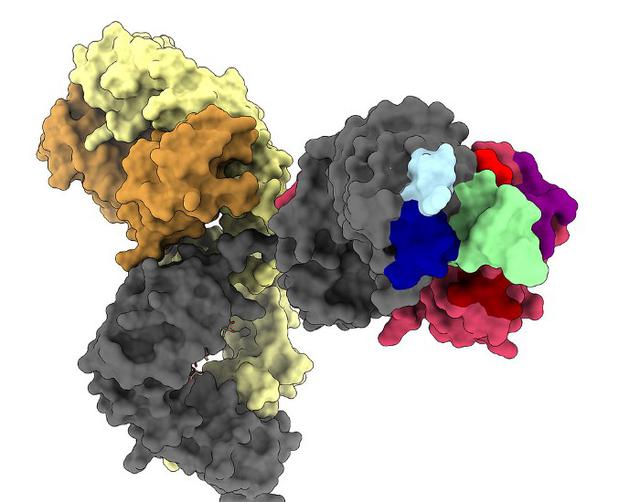
So I've submitted the thesis, had the viva, gotten the result and it's the end of a two year stint at trying bioinformatics. How did it go? Well, quite well I think but there are definitely some key learning points and outcomes that I feel are worth sharing.
I've uploaded all the code and methodology to my github page so you can get a better look at what some neural networks look like (hint - pretty messy I'll admit but we can improve in the future!).

Largely, in machine learning, it's all about the data. This was stressed at the beginning but I still didn't do enough. In biology, data is extremely dirty! Other fields have data issues too, sure, but not like biologists. Living systems are complicated, enormous and difficult to capture at high accuracy. In our case, structures probably had mistakes within them.
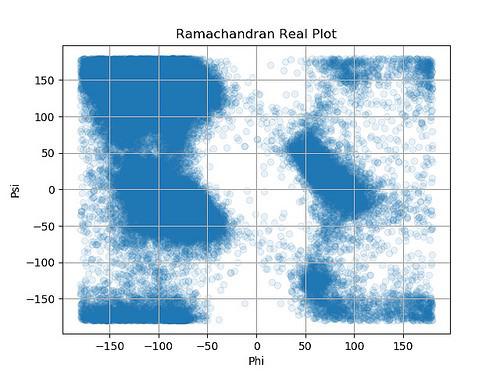
The plot above is called the Ramachandran plot and it's used quite a bit in structural biology. It shows that certain combinations of the phi and psi angles are much more common than others. Had I looked at this graph much earlier, alarm bells would have gone off - we probably had a lot of erroneous models in there and we perhaps should have cut them out.
Redundancy was another big problem. Quite often, the same model might appear multiple times in a set, under another name. Maybe it's the same structure with a different resolution? Maybe it was thought to be different at the time but was actually the same thing. When you are dealing with neural networks you don't want them to start remembering what they've seen by showing the same datum over and over again.
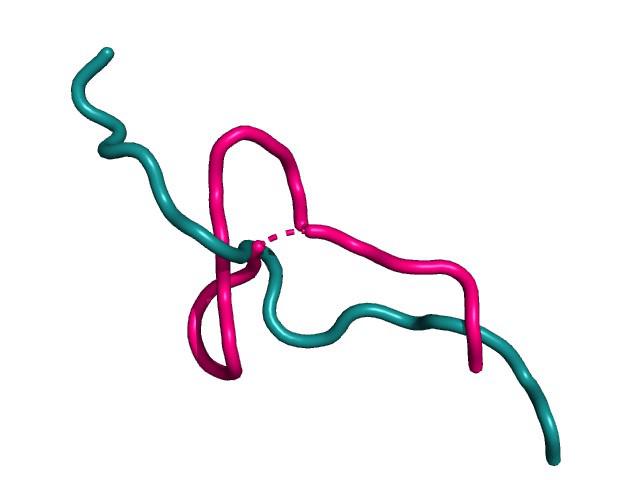
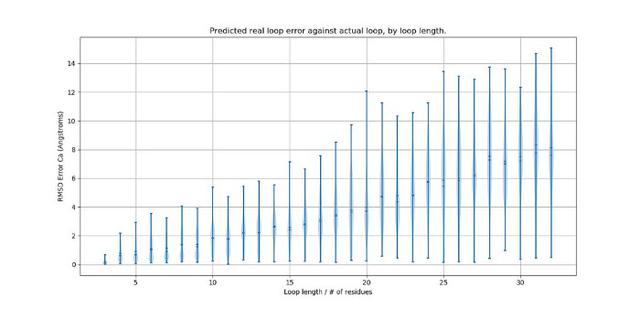
Early on, there was a question about how flexible these loops are, and how that might affect the modelling. I looked around a bit but never found a fully satisfactory answer. I left it and carried on regardless. It came back to bite me, or at the least, a little nibble. It still annoys me that I never really found out and perhaps that's where the real science actually lies? If you can't find the answer in the literature, maybe it's because it's not actually been looked at, and that's exactly where you need to be. It might be a little scary but that's where the thrill is too. It's a gut-feeling in someways and one shouldn't ignore it.
This is key really! Science is built on the idea that an experiment can be reproduced and if followed correctly, should yield the same result. When it comes to this sort of work, having clear, concise code, with a path from data all the way to results is very important. My thesis contains all the pertinent code but all the experimental python lives in a github repository, online where anyone can get it.
Zenodo is an attempt to store code and datasets in order to make projects more reproducible. I'll be placing my LoopDB database there very soon. The code I've used can also be found there, along with instructions on how to train your own networks.
Oh, and don't forget to mention or cite your friendly High Performance Computing (HPC) people :)
It turned out that we couldn't reject the null hypothesis; we didn't manage to improve on the state-of-the-art. Our models didn't really fit quite right and lacked the accuracy we needed. This is perfectly fine in science even if it is a little depressing. There is a bit of a problem in science when it comes to publishing papers that have negative results - it's been spoken about extensively. We are all human though and it's understandable to feel a little down when things don't go as planned. But this is an opportunity and a sign to change tack and try other things.
This is what we did when we looked at scoring and the results got a little more interesting.
This is more of an engineering or artisan aspect but I think it's worth serious thought. One thing we did get right was to convert all our PDB files to entries in a postgresql and mongodb database (mongodb seems less bad than it used to be I'd hasten to add). This way, generating subsets, particular loops and filtering was quite fast and easy enough to backup.
Where it went a little wrong was using Python for everything. While Python is pretty much the go-to for tensorflow, using something like the R language for analysing the data might have been a lot faster. Get python to spit out data that R can easily spin around, plot, filter and digest. Maybe moving the best neural network to C and running it that way would have been even better? A good mix of tools and using the right one for the right job is an underrated skill I think.
Of course, git is essential in any software development, but getting a backup and versioning strategy sorted early on is a real plus. One thing I was fortunate enough to be able to do is build a second PC with a GPU, from spare parts that were being thrown away. This box could run all the experiments whilst I designed and tested new nets on the main machine. Keeping track of all these running experiments with trello seemed to work quite well, especially when we began to use cloud services, in addition to my home box.

Zotero is essential for anyone doing this sort of thing. Honestly, I don't know why anyone would use anything else if I'm honest. It's free, exports to all major bibliography formats, has an excellent Android client you can use to read your papers in a comfy chair, and is easy to use. Forget Mendeley, Endnote and all that crap. And no, I don't work for Zotero :P
Using a time tracking application on my second machine helped too! It can be easy, if you are me, to lose track of time or overestimate the amount of time you've spent. I know a few freelance folks who keep track of time with pomodoro affairs and what not but I use a program that allows me to keep time spent against particular projects.
Finally, turning off email, twitter, facebook or anything like that is a must! Keeping such distractions out however you can seemed to help me.
One of the problems is I'm a computer scientist coming at the biological sciences with no prior experience! It can be all too easy to fall back on what you already know and the ways you do things. Speaking to folks and talking about your research with others is a really valuable thing to do.
These communities might not always be in your lab or even your university or subject. I found that the London Biohackspace was really helpful in understanding a couple of algorithms I couldn't quite get my head around.
When you are working remotely this can be really tricky and it took it's toll on me I'd say. If I did this again, I'd probably seek out some more local groups I could go and visit to talk about my work. I did attempt to talk at a conference about my work but it came at a really bad time and I had to pull out. It's a shame really, but it's definitely a good idea to speak to other professionals when you can, even if it's not something you enjoy doing. Even a little will help.
Setting up a routine with your supervisor is very important. Despite the distance and timezone difference I was upfront with my supervisor and we spoke every two weeks without fail. Getting a list of questions and thoughts before your meeting is a good idea as supervisor time is quite valuable.
One of the tools I didn't mention was my own wiki. I kept a diary throughout the project which came in useful towards the end. I saved a whole 4 months for writing and finishing, which is 33% of the total time. It sounds like a lot but I highly recommend it! I think, on reflection I'd have written parts of chapters as I went along.
People have recommended scrivener to me and I've tried it once. It's pretty good but for this project I went full on Latex. Going forward I'd suggest a combination of both somehow, where one can organise one's thoughts using scrivener first, then Latex just for layout. Sadly, scrivener doesn't have a Linux client. One can get someway there with include files within latex but it is a little clunky. I'm open to alternatives on this one.
Doodling and keeping notes in some form of notebook is a good thing, but I did find that I never really referred to them that often. I think perhaps some mind-mapping software might be called for in the future. Keeping everything in your head all at once is impossible. Never overlook a good whiteboard, an empty wall and a large stack of post-it notes!
In the end, I spent a lot of time talking to different supervisors, trying to get a good balance between a project I liked, what the supervisor relationship might be like, and what the outcomes might be. I believe I chose quite well but I did have to do a lot of leg work to make that happen before anything got started. I'd say this is the number one tip. You do need to enjoy your project and that can be really hard to assess before you've even started but speaking to folks in person will get you quite far.
Birkbeck is a good university if you find yourself wanting to change tack and study whilst working. Aside from this, the structural biology group is very good and if you decide to go down a similar route, I can't recommend them enough.
You'll have down times, you have stresses and it won't be easy at all. It can be a lonely place but there are moments that are just glorious! It's an adventure!
